Gene Conservation Laboratory
Microsatellite Analysis and Sequencing
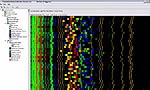

Analysis of microsatellite DNA is done on an automated capillary sequencer. Each of the 48 horizontal lanes in this photo represents a single fish migrating through a single capillary. Fluorescent chemistry allows the detection of five different genes in each lane of this multiplex run. One advantage of microsatellite analysis is that tissue samples do not need to be frozen. In microsatellite analysis, the scoring of genetic variation is made easier by software that teases apart the individual loci resolved using the fluorescent detection system.

The PCR samples are prepared for electrophoresis by adding diformamide, to assist in denaturing, and an internal lane size standard. An automated Biomek® FX processes samples in 384-well plates for high throughput and reduced loading errors.

After the amplified samples are loaded and the plates sealed to prevent evaporation, they are denatured at 95C, cooled, and contained in plate assemblies for loading on the ABI 3730 Capillary Sequencer.


Preparing the 3730 for Microsatellite analysis. New run buffers, rinse water, and waste water must be made after 96 microsatellite runs or 48 sequencing runs. The electrophoretic gel must be replaced weekly.
Plate assemblies are loaded into the 3730 and a run is initiated.

The sequencer automatically analyzes DNA molecules labeled with multiple fluorescent dyes. After samples are automatically loaded into the system's capillary array, they undergo electrophoresis, laser detection, and computer analysis. Electrophoretic separation can be viewed on-screen in real-time, and final data can be output in a variety of formats.

After electrophoresis, the run files can be transferred to another computer work station ready for analysis.

Analysis of sequencing DNA is also performed on the ABI 3730 Capillary Sequencer. The 24 horizontal lanes in this photo of a real-time read represent a single fish migrating through a single capillary. The bottom 24 are the reverse complement of the top fish. ABI BigDye® v3.1 Cycle Sequencing chemistry allows for detection of the four nucleotides. Preparing sequencing samples to run on the 3730 is the same process as Microsatellites with one additional step immediately after PCR amplification.

Unincorporated Dye Terminators must be removed from the PCR reaction prior to loading on the 3730 for accurate detection of sequence. Qiagen DyeEx Kits are used for this procedure.
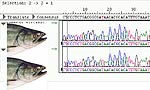
After electrophoresis, the run files can be transferred to another computer work station and imported into sophisticated sequence analysis software.
