Gene Conservation Laboratory
Alaska Peninsula Chinook Project

In the 1990s, ADF&G contributed to the development of a genetic baseline for Chinook salmon (King salmon; scientific name: Oncorhynchus tshawytscha) using protein variation to infer genetic information about salmon populations (Crane et al. 2000,Teel et al. 1999). Record low runs of Chinook salmon in Alaska resulted in increased interest in developing a more comprehensive genetic baseline, a key component for performing mixed stock analysis (MSA) of fisheries using genetic data. At this time the Gene Conservation Laboratory (GCL) was continuing to develop baselines for use within river systems (Yukon River — Smith et al. 2005; Kuskokwim River — Templin et al. 2011; and Copper River — Seeb et al. 2009 ) using newer, DNA-based genetic markers, especially single nucleotide polymorphisms (SNPs). This foundational work led to the GCL completing the first comprehensive genetic baseline for Alaska Chinook salmon and surrounding regions (Templin et al. 2011) which allowed extension of genetic analyses to marine waters (e.g. juvenile studies - Murphy et al. 2009; migration — Larson et al. 2013; and fishery bycatch — Templin et al. 2011; NMFS 2009; Guyon et al. 2010 a,b).
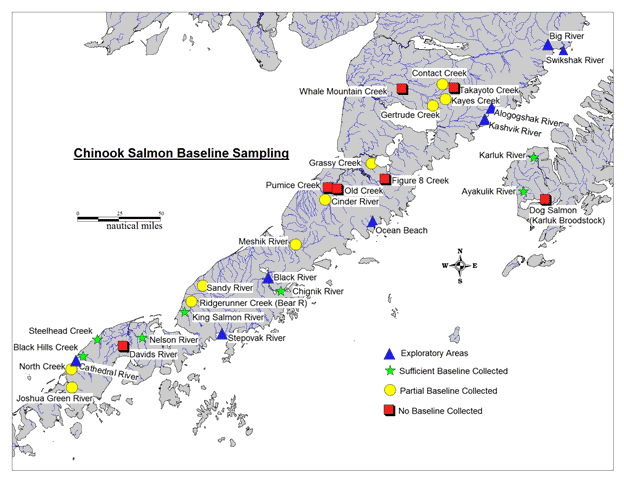
The more comprehensive genetic baseline allowed for examination of potential impacts of marine fisheries on Chinook salmon stocks. Recent studies have revealed that a larger than expected percentage (14-27%) of Chinook salmon from the north Alaska Peninsula are caught incidentally in the Bering Sea Pollock fishery (NMFS 2009; Guyon et al. 2010 a,b; Guthrie et al. 2012). While the north Alaska Peninsula populations were adequately represented within the existing baseline for large-scale analyses, there were several stocks in the Westward Region that did not have good representation in the genetic baseline, specifically on the Alaska Peninsula and Kodiak Island (see Figure 1 below).
This project was designed to obtain samples of known Chinook salmon stocks to expand the genetic baseline and to investigate the existence of additional stocks that are not currently known by the Department. This intensive survey and sampling of Chinook salmon spawning locations on the Alaska Peninsula and Kodiak Island will provide better knowledge of the distribution, size and biology of Chinook salmon populations in this remote area.
Some of the Chinook salmon production on Kodiak Island comes from Pillar Creek Hatchery which used Karluk River as the source of its original broodstock. We collected samples of the hatchery broodstock as part of this project that, when combined with samples of their progeny, provide an opportunity to assess the potential for parentage-based tagging (PBT; Anderson and Garza 2006) of the hatchery production in tandem with traditional MSA. If successful, PBT will allow identification of hatchery offspring in all fisheries where Pillar Creek Chinook salmon are harvested. The potential for this application depends on the ability of sets of single nucleotide polymorphisms (SNPs) to identify offspring based upon parental genotypes and known mating pairs. The GCL is currently investigating a large suite of SNPs for such applications. The combination of the improved genetic baseline from the Westward region for MSA purposes and the potential for PBT applications may provide powerful new genetic tools to improve our understanding of Chinook salmon populations in the region.
Other Works Cited
- Guthrie, C. M. III, H. T. Nguyen, and J. R. Guyon. 2012. Genetic stock composition analysis of chinook salmon bycatch samples from the 2010 Bering Sea trawl fisheries. U.S. Dep. Commer., NOAA Tech. Memo. NMFS-AFSC-232, 22 p.
- Guyon, J. R., Guthrie, C. M., and H. Nguyen. 2010a. Genetic stock composition analysis of Chinook salmon bycatch samples from the 2008 Bering Sea pollock fishery. Report to the North Pacific Fishery Management Council. pp. 32.
- Guyon, J. R., Guthrie, C. M., and H. Nguyen. 2010b. Genetic Stock Composition Analysis of Chinook Salmon Bycatch Samples from the 2007 "B" Season and 2009 Bering Sea Trawl Fisheries, Report to the North Pacific Fisheries Management Council. pp. 32.
- North Pacific Fishery Management Council, 2009. Bering Sea Chinook Salmon Bycatch Management, Final Environmental Impact Statement, printed by National Oceanic and Atmospheric Administration, National Marine Fisheries Service, Alaska Region, Juneau, Alaska.
- Anderson, E. C. and J. C. Garza. 2006. The power of single-nucleotide polymorphisms for large-scale parentage inference. Genetics. 172(4): 2567-2582.